Dell Medical School, UT Austin
Apricity Health
egulators and drug developers are intensely focused on boosting diversity among clinical trial participants, in part because so many of the new precision oncology drugs prove less effective in real-world populations than they do in highly selected clinical trials. Many factors contribute to this loss of effectiveness in real-world care, and increasing diversity in clinical trial participation alone is necessary but insufficient.
A key piece of data drug developers rely on to make decisions about which targets to develop drugs against is cancer genomic data. However, about 85 percent of US cancer patients are diagnosed and treated outside of major academic institutions where most of these types of genomic studies are conducted, and therefore they are infrequently engaged in such studies. As a result, the clinical, genomic, and therapeutic response data from this 85 percent are under-represented in research data sets, rendering these patient subpopulations invisible to the drug development engine.
This is a lose-lose reality for patients and drug developers. When newly approved drugs are prescribed to the broader real-world populations, they are less likely to be effective in patients who have not been studied. Conversely, biopharma could be missing out on untapped therapeutic opportunities unique to these under-represented populations. The result is not only missed opportunities but also loss of precious time for patients.
Clinical Trial Diversity is Necessary but Insufficient
While these efforts must continue, we bring attention here to a less-talked-about but equally profound structural bias in the system, namely the lack of inclusiveness and representation in foundational genomic data sets.
Each patient’s tumor is different. Advances in sequencing technologies have afforded us the ability to sequence the DNA in each patient’s tumor and unequivocally demonstrated that each patient’s tumor is unique despite common mutations. Furthermore, the presence or prevalence of a particular driver mutation can vary not only across tumor types but also ethnic or racial populations, even when they share similar histopathology under the microscope. Since cancer genomic data has become the de facto starting point, or at least the key piece of data, in the process of identifying and prioritizing a target for therapeutic and/or biomarker development, it stands to reason that under-representation of ethnic or racial populations in such foundational research data sets can instigate systematic bias that is propagated down the drug discovery and development pipeline.
Representation Bias in Research Genomic Data
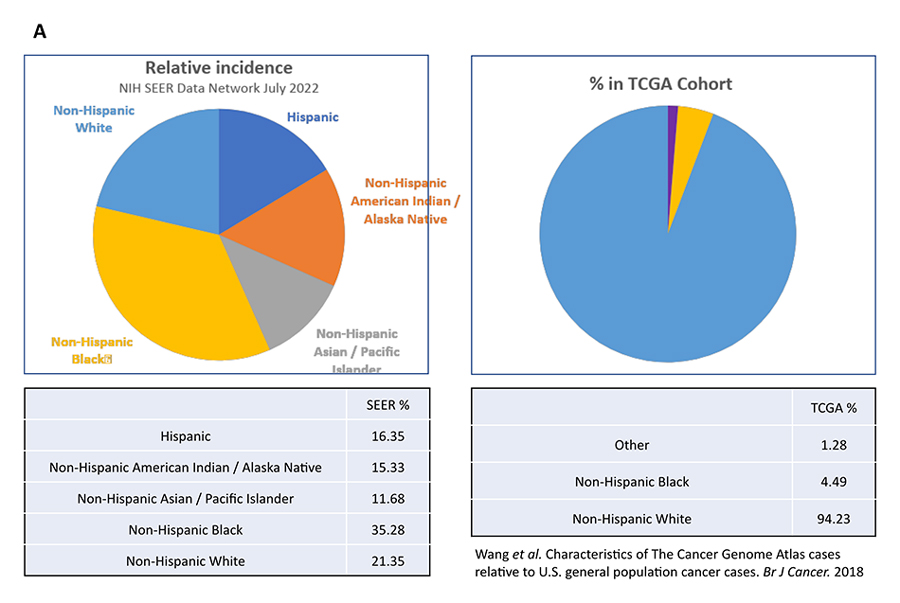
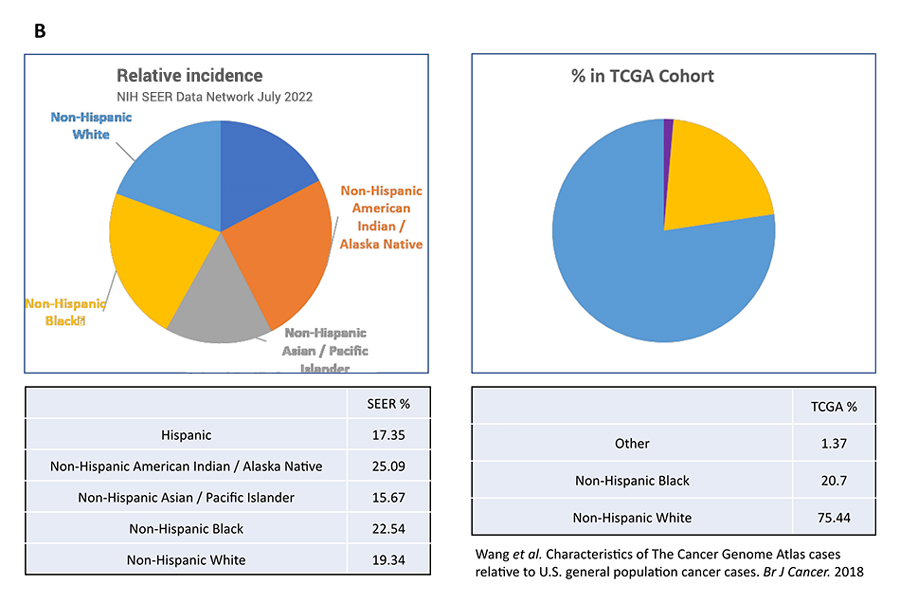
However, when we compared the ethnic and racial make-up of the patient populations studied in TCGA with SEER (Surveillance, Epidemiology, and End Results Program) age-adjusted incidence rates for a cancer type, significant imbalance exists. For example, while only 21 percent of US prostate cancer patients are identified as non-Hispanic White, 94 percent of the prostate tumor samples in the TCGA were collected from non-Hispanic White patients (Fig. 1A). That means 79 percent of the prostate cancer patient population is under-represented in TCGA. This is not a one-off observation. As shown in Figure 1B, 75 percent of the colon tumor samples in TCGA came from non-Hispanic White patients, which account for only 19 percent of US colon cancer patients. In other words, the genomic landscapes of colon cancers in Hispanic, Asian, and Native American patients are not studied and understood at the same depth as the same histopathology in Whites.
Therefore, in order to ensure equitable access to effective anticancer treatments, we must innovate solutions to tackle the origin of structural bias in the therapeutic R&D pipeline.
Embed Genomic Research in Everyday Routine Care to Overcome Representation Bias
Most deep multidimensional genomic studies are being conducted in major academic centers, where resources, capabilities, and commitment exist for both the study team and the patients. However, this will always be limited to a small subset of the total patient population, especially for the 85 percent who are treated outside of these major centers. In the real world, particularly in rural communities, it is neither feasible nor practical to enroll patients in similar kinds of studies given the participation burden on patients who are already facing many access barriers.
For example, serial tumor biopsies produce crucial biospecimens for analyzing and understanding how resistance emerges on therapies. However, participating in such a research study requires multiple visits to research sites that may be far away, potentially undergoing invasive biopsy procedures, among other requirements. Many patients would find it challenging to participate in such a study. Instead, a study designed to collect serial liquid biopsy samples in conjunction with routine clinical lab draws would be easy for many more patients, because every patient in routine care will be monitored with routine labs. Consenting to an extra tube of blood for research with the same needle stick represents a negligible burden to participate. While such liquid biopsy samples would not generate the same depth of data as tumor specimens, they will provide a broader view across a more diverse population of patients, which is necessary for evaluating the general relevance of insights from deep molecular studies of tumors obtained from a narrow patient cohort. In other words, these studies are complementary.
We cannot simply export the same study designs from academic centers to rural or other communities. We need to reimagine how and what kind of genomic research can be embedded into everyday routine care, so it is easier for more patients to participate. Such patient-centric studies may have to be more focused, compared to typically comprehensive deep research at academic centers.
In summary, representation bias in research genomic data sets can lead to structural bias that is propagated downstream along the entire drug discovery and clinical development pipeline. Equitable access to effective anticancer treatment, therefore, must begin with inclusion of diverse representative populations in foundational research studies, so that precision drugs can be developed that work for every patient to whom they are uniquely suited.

