Ellison Institute
@ellisoninst
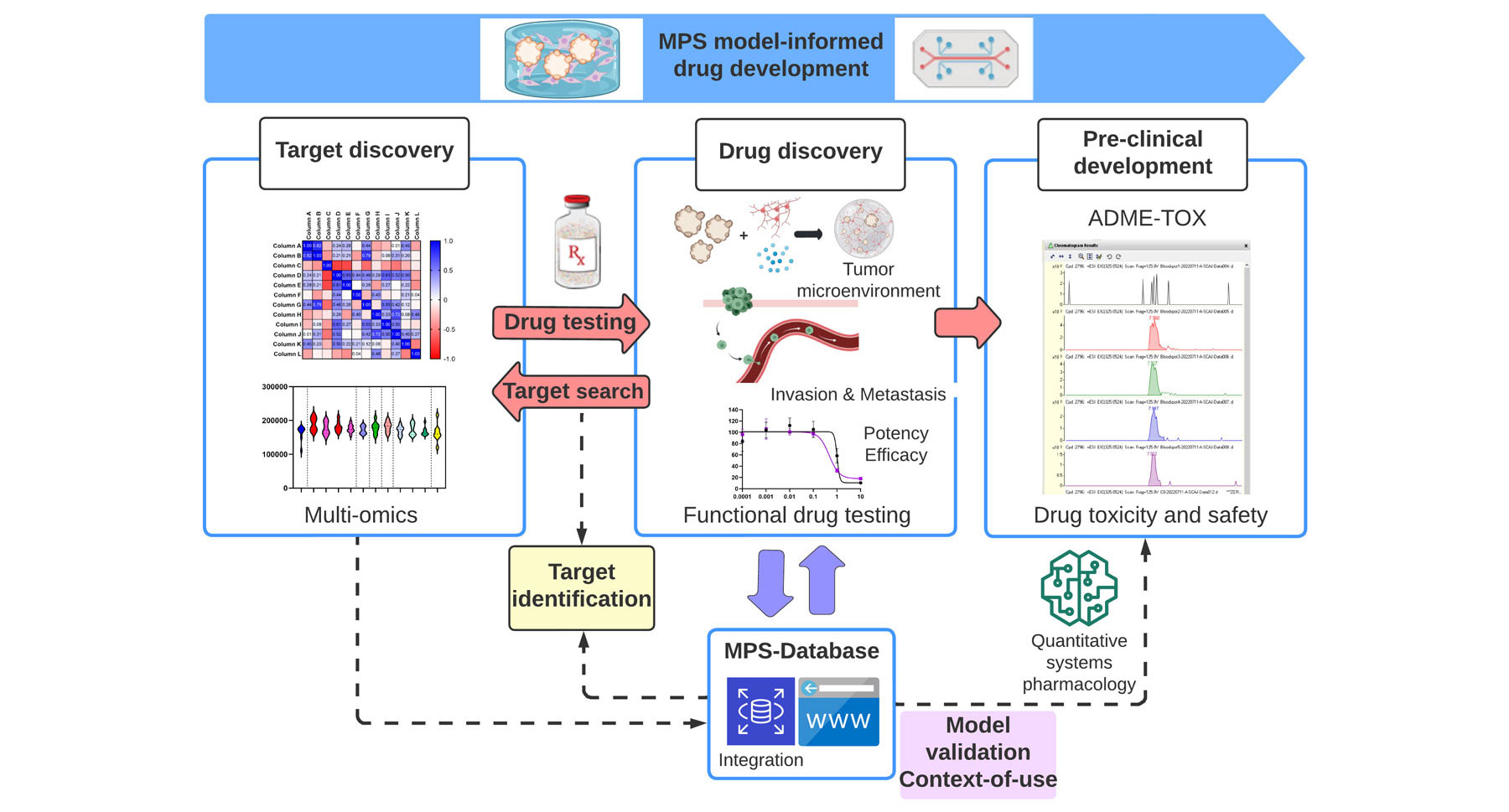
icrophysiological systems (MPS) are platforms for in vitro modeling of a specific tissue or organ by exposing cells to a microenvironment that mimics the physiological aspects important for their function. Coupled with functional drug testing, MPS could contribute to the ongoing transformation of the drug-development process by recapitulating organ-level pathophysiology and clinical response. A major challenge lies in integrating multiscale, context-specific data to demonstrate the utility of these emerging technologies and establish context-of-use. Here, we highlight the current use of MPS for drug safety testing, suggest expansion to discovery and pre-clinical development of oncology drugs, and advocate for systematic data collection and data sharing for successful integration.
The Drug Development Crisis
A First Step Towards Improvement: Functional Drug Testing
Over the past two decades, efforts such as The Cancer Genome Atlas (TCGA) have provided a foundation for researchers to apply functional genomics workflows to turn molecular alterations found in treatment-naïve patient tumor samples into potential candidate targets for new pre-clinical campaigns. Recognizing the need to extend best practices of data generation and sharing from TCGA, the Cancer Target Discovery and Development (CTD2) network was established in 2009 and has served as a resource to share functional genomics workflows and research results using a tiered level of evidence following FAIR (Findable, Accessible, Interoperable, and Reusable) principles.
Many researchers have benefited to date, including those at the Ellison Institute, by using these resources to compare findings and avoid duplicating pre-clinical efforts. However, results from the largest precision oncology trial to date, the National Cancer Institute’s Molecular Analysis for Therapy Choice (NCI-MATCH), estimate that only 5 percent of patients with actionable alterations will benefit. These results demonstrate that the current paradigm of relying solely on static snapshots from patient samples is unsustainable. Efforts connecting spatio-temporal data from animal models to human phenotypes have been successful to support craniofacial research (e.g., facebase.org), which cancer initiatives such as the Human Tumor Cell Atlas could build upon. The FDA’s commitment to advancing model-informed drug development (MIDD), codified in the 2017 Prescription Drug User Fee Act Reauthorization (PDUFA VI, 2018–2022), is underscored by the program’s continuation as part of PDUFA VII. Moreover, the past decade has led to a notable increase in the number of regulatory submissions that contain quantitative systems pharmacology (QSP), an integral part of MIDD. In line with the goals of MIDD, multiscale data sets can support powerful new insights and decrease uncertainty at key decision points in the development process.
Finding Functional Targets Using MPS
Cancer drugs, including those in late-phase clinical trials, often kill cancer cells through off-target toxicity, highlighting a need for improved target validation. High-affinity, target-specific binding doesn’t always translate to effective inhibition. In fact, we recently discovered a mechanism by which small molecules promote the oncogenic signaling that they were designed to block, highlighting the need for functional and bidirectional assays in pre-clinical testing. Historically, target validation and pre-clinical testing have been performed using immortalized cell lines grown in a 2D environment in the absence of immune components and other aspects of the tumor microenvironment, including cancer-associated fibroblasts (CAFs), endothelial cells, the extracellular matrix, and physiologically relevant mechanical forces. This simplified approach also fails to recapitulate interpatient heterogeneity and is therefore not amenable to precision medicine.
Over the past 10 years, researchers have developed a range of MPS technologies for more accurate representation of human physiology and better prediction of drug response. For example, several studies have shown that patient-derived organoids mimic and even predict patient drug response. They can be readily scaled up for high-throughput screening, and more complex co-cultures with CAFs and immune cells are also surfacing. Organs-on-chips are engineered to yield functional tissues capable of modeling organ-level responses. They recapitulate tissue-to-tissue interfaces, use microfluidic channels to generate shear stress, and often provide mechano-actuation to emulate breathing (lung) or peristalsis (intestine).
Industry and regulatory agencies recognize the promise of MPS but understand that they must be extensively characterized, tested, and cross-validated to understand their potential limitations, value, and individual context-of-use. The specific role of MPS in pre-clinical testing has yet to be fully realized, as they could be used alongside animal models to assess species-specific drug toxicities and/or perform efficacy testing, and eventually reduce or even replace animal models. Organizations such as the Health and Environmental Sciences Institute (HESI) and the National Centre for Replacement, Refinement, and Reduction of Animals in Research (NC3R) are working with the FDA to promote the development and application of this technology, to facilitate its expanded scientific and regulatory use.
Call to Action: Integrating Multi-Scale, Context-Specific Data for Context-of-Use
Ideally, we could start with a high-quality data resource that aids in study design, experimental setup, analysis, visualization, and computational modeling, all with secured access. Biosystics-AP™, formerly known as the University of Pittsburgh Microphysiological Systems Database (MPS-DB) and one of the three NCATS-funded TCTCs, aims to provide such a resource. It supports aggregation, analyses, and computational modeling to facilitate studies of disease mechanisms, drug and compound toxicity, and PK/PD drug predictions.