Chief Clinical Research Officer
Dana-Farber Cancer Institute
@DanaFarber
Professor of Medicine
Harvard Medical School
@harvardmed
he genomic and immunohistochemical characterization of non-small cell lung cancers (NSCLC) and the consequential development of effective targeted and immunotherapy has provided significant ongoing incremental progress in treatment of this deadly cancer. This article outlines this progress, confirming the value of biomarkers and the assays that measure them to facilitate drug development and make precision medicine a reality for nearly half of lung cancer patients.
Genomic Changes with FDA Approved Agents for Patients with Lung Cancer
Agents Active in Patients with Acquired Resistance to First Generation Targeted Agents Yield Long Progression-Free Survival When Used as Initial Therapy
EGFR Mutant Lung Cancer: Research findings from studying patients with the two most commonly targeted oncogenic drivers, EGFR and ALK, have yielded more effective anticancer compounds. The first two EGFR-TKIs approved for patients with lung cancer and sensitizing EGFR mutations were gefitinib and erlotinib, first-generation EGFR inhibitors. Patients whose EGFR-mutant lung cancer developed resistance to these first-generation EGFR-TKIs were systemically biopsied and sequenced, revealing that most of the patients acquired a T790M mutation within the EGFR gene which mediated the resistance to EGFR-TKIs. Preclinical experiments showed pyrimidine-based compounds rather than the previously deployed quinazalone-based compounds such as gefitinib, erlotinib, and afatinib, were active in cell lines and animal model systems with both a sensitizing mutation (L858R or exon 19 deletion mutations of EGFR) and the acquired resistance mutation T790M. Osimertinib is one of the pyridmidine-based compounds and was initially approved in 2015 for patients with sensitizing mutations of EGFR and resistance to EGFR-TKIs mediated by an acquired T790M mutation. The agent osimerinib was then studied as initial therapy for lung cancer patients with sensitizing mutations of EGFR. The lung cancer patients with sensitizing mutations of EGFR treated with the more effective osimertinib have a progression-free survival of nearly twice as long (19 months versus ten months) compared to patients treated with the first-generation agents erlotinib and gefitinib.
ALK Rearranged Lung Cancer: A similar story has happened with lung cancer patients with ALK rearrangements. Crizotinib was the first ALK inhibitor approved as initial therapy for lung cancer patients with ALK rearrangements. Patients whose ALK-rearranged lung cancers developed resistance to the first-generation ALK inhibitor crizotinib were systemically biopsied—revealing a myriad of resistance mechanisms. A more potent and specific inhibitor of ALK, alectinib, was active against many of these mechanisms of resistance and caused responses in the majority of ALK positive patients with acquired resistance to crizotinib. Alectinib was approved in 2015 for lung cancer patients with ALK rearrangements with acquired resistance to crizotinib. The agent alectinib has now been deployed as the initial therapy for lung cancer patients with ALK rearrangements. Lung cancer patients with ALK rearrangements treated with the more effective alectinib had a progression-free survival of nearly three times as long (35 months versus 11 months) compared to patients treated with the first-generation agent crizotinib; alectinib is now the agent of choice as initial therapy for ALK positive NSCLC.
Precision medicine has been effectively deployed in lung cancer for ALK rearrangements and EGFR sensitizing mutations. Genomics studies provide insights into drug development by identifying the appropriate genomic changes that can be effectively treated with the targeted agent, and provide insights when resistance develops to define mechanisms of resistance and develop drugs to overcome that resistance. The paradigm of deploying osimertinib and alectinib as newer and more effective agents for initial therapy has been an effective strategy for EGFR mutations and ALK rearrangements respectively.
Precision Medicine Defining Subsets of Patients with Lung Cancer Who Are Likely to Benefit from Checkpoint Inhibitors
Immunotherapy for Patients with EGFR mutations and ALK rearrangements: There are currently four different checkpoint inhibitors approved for different therapeutic indications for lung cancer: nivolumab, pembrolizumab, atezolizumab, and durvalumab. Thus far, cancer genomics have been less helpful in identifying patients likely to benefit from immunotherapy than from targeted therapy. In addition, clinical research has shown there is little overlap between the patients who benefit from targeted therapy with EGFR and ALK inhibitors and those that benefit from checkpoint inhibitors. It is rare for a patient with an EGFR mutation or ALK rearrangement to respond to a checkpoint inhibitor, so patients with somatic EGFR mutations and ALK rearrangements have typically been excluded from checkpoint inhibitor registration trials.
PD-L1 Testing: The leading predictive marker used to identify patients more likely to benefit from checkpoint inhibitors is immunohistochemical staining of the tumor for program death ligand 1 (PD-L1). The reproducibility of the PD-L1 immunohistochemical analyses across trials for these four different checkpoints has been complicated by the use of different PD-L1 assays with different antibodies, different cut-points, and different approaches to PD-L1 staining (tumor cells alone versus tumor cells plus other non-malignant cells making up the tumor). Despite PD-L1 being studied in all the registration trials for these four agents, only pembrolizumab requires the presence of PD-L1 for its indication, needing one percent of the tumor cells for previously treated non-small cell lung cancer, and 50 percent of the tumor cells as a single agent, first-line therapy for non-small cell lung cancer. Pembrolizumab and atezolizumab are also FDA approved for use as first line therapy with chemotherapy without PD-L1 testing for non-small cell lung cancer.
Despite selecting the non-small cell lung cancer patients with the high PD-L1 staining of more than 50 percent (30 percent of screened patients), the response rate to pembrolizumab was still below 50 percent, a response rate less than the 60-80 percent response rates typically seen with patients with EGFR mutations or ALK rearrangements treated with targeted agents. In a similar trial design for patients with non-small cell lung cancer, patients were treated with either nivolumab or chemotherapy if their tumor had five percent or more of their tumor cells staining for PD-L1 (40 percent of screened patients). The response rate for those treated with nivolumab alone in the target population was about 25 percent. The response rate and other outcomes of the trial did not lead to FDA approval of nivolumab in patients with five percent or more of their tumor cells staining with PD-L1 antibody. The field urgently needs some standardization of the PD-L1 assays and some agreement on cut-points that can be implemented in clinical trials in different settings, so that findings across trials can be compared.
Tumor Mutation Burden: A measure of genomic changes determined by DNA sequencing has been added as a potential predictor of benefit using whole exome sequencing and next generation sequencing panels. The tumor mutation burden has been described by the number of somatic mutations per megabase of DNA. The field has thus far been hampered by different sequencing methods (whole exome or different next generation sequencing platforms); a minority of patients having adequate tumor specimens available for genomic characterization; different cut-points in mutations per megabase; and lack of prospectively defined entry criteria of mutation burden for clinical trial entry. The response rate of previously untreated non-small cell lung cancer with a high mutation burden (more than 10 per megabase) treated with nivolumab still remains below 50 percent.
Human Tumor Atlas Network: The response rate to treatment with single agent checkpoint inhibitors in unselected patients with lung cancer and most other solid tumors remains below 20 percent. In addition, there are rare patients with EGFR mutations and ALK rearrangements who do not benefit from targeted therapies. The reasons for the lack of response remain incompletely defined, and the leading predictive immunohistochemical and genomic biomarkers are thus far incomplete. Defining the immune and cellular microenvironment in addition to the tumor cells present in human tumors offers the opportunity to further characterize the determinants of response and resistance to checkpoint inhibition in some solid tumors. A Human Tumor Atlas Network (HTAN) has just been formed to address these issues as part of the Beau Biden Cancer MoonshotSM Initiative within the NCI. The goal of the HTAN is the construction of human tumor atlases that describe the multidimensional cellular, morphological, and molecular mapping of human cancers over time for informing future cancer research and, ultimately, making clinical decisions. The ongoing genomic studies and the mapping of human tumors will likely add to our ability to predict who is likely to respond; to understand the reasons for acquired resistance; and to inform the selection of appropriate therapeutic interventions that will modify the tumor microenvironment and increase immunotherapy efficacy in common human tumors including melanoma, colon cancer, lung cancer, breast cancer, and pancreatic cancer–all solid tumors within the Human Tumor Atlas Network. We remain hopeful the ongoing genomic, immunohistochemical, and spatial analyses will continue to inform the predictive markers of sensitivity and resistance to both immunotherapy and targeted therapy and allow appropriate therapeutic interventions to improve the outcome of patients with lung cancer and other solid tumors.
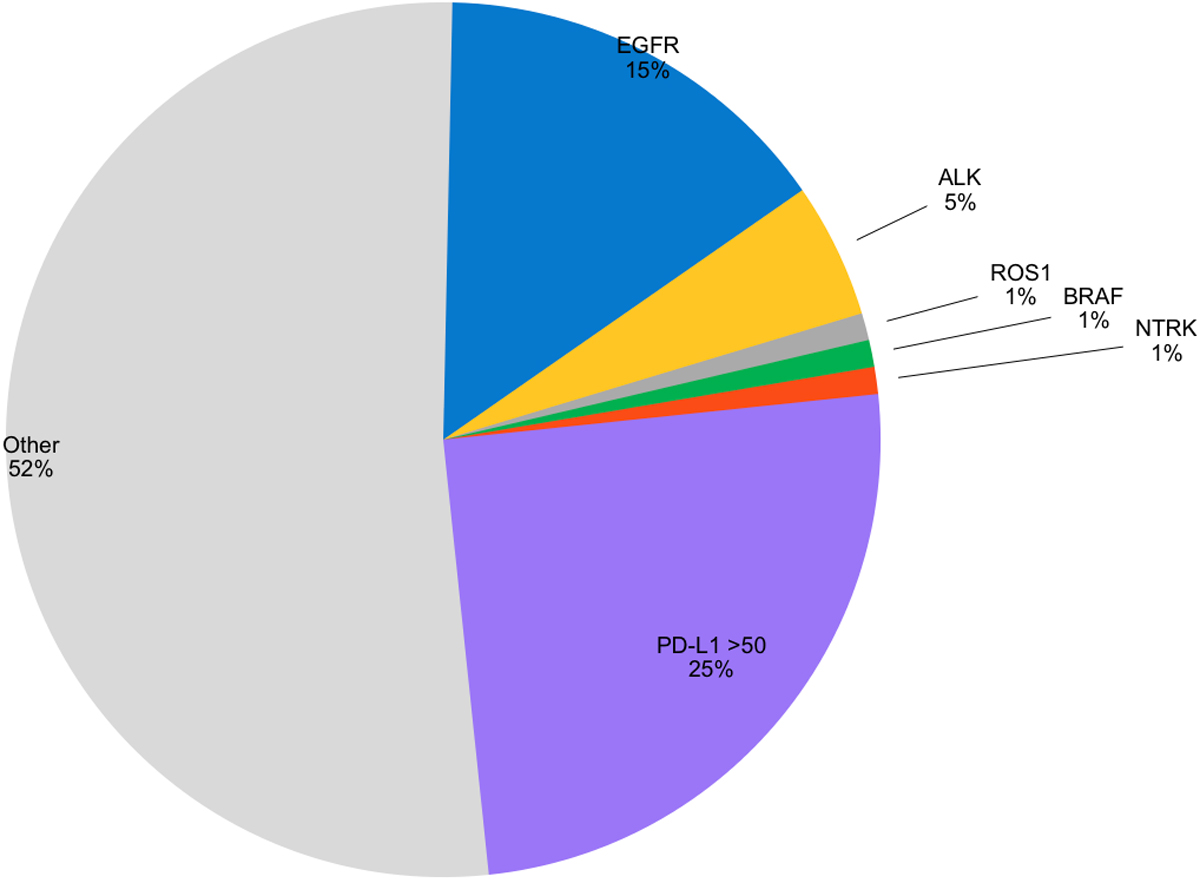
Figure 1: FDA-approved targeted agents are available for 5 genomic changes (EGFR, ALK, ROS1, BRAF, NTRK), accounting for 23% of patients with adenocarcinoma of the lung. PD-L1 testing provides a predictive marker to identify patients likely to benefit from single agent checkpoint inhibitors.

