Improving Imaging Biomarkers in Oncology
Lawrence H. Schwartz
Tavis Allison
Binsheng Zhao
his month’s Translational Research discussion features imaging biomarkers, their essential role in predicting clinical trial outcomes, and new approaches which may accelerate drug development and protect patient safety.
This article summarizes progress from the initial imaging biomarkers developed to evaluate tumor shrinkage in response to cancer therapeutics, to the challenges posed by contemporary treatment paradigms which may produce new patterns of response. Current avenues toward meeting this challenge, including volumetrics, radiomics, kinetic modeling, and artificial intelligence, are reviewed. It focuses on computerized tomography (CT) because it is the workhorse of contemporary oncology, generating datasets required to develop novel approaches and creating immediate translational potential. However, the techniques discussed may enhance analysis of any quantitative imaging modality, including FDG-PET and/or MRI. The review of historical progress suggests factors that drive improvement independent of technology: increasing reproducibility, refining conceptual models, quantifying tumor phenotype, and learning from experience.
RECIST: Reproducible Quantification Within a Categorical Conceptual Model
Biomarkers in oncology developed because the growing number of potential therapeutics demanded comparison of response between clinical trials at different institutions. The World Health Organization (WHO) addressed this need in 1981 through consistent guidelines for progression and response. Tumor size defined the categories of partial response (PR) and progressive disease (PD), because shrinkage historically predicted survival with cytotoxic therapies and radiation treatments. Because then-contemporary medical imaging could only localize and visualize tumors, quantitative measurements were made with rulers or calipers. The measurement error of this technique was estimated by comparing measurements of phantom spheres under a foam sheet, leading the WHO to set PR’s cut-off at 50% decrease in the bidimensional measurement. From 1994 to 2000, a large international collaboration updated these WHO guidelines. After reviewing thousands of cases and finding no differences between the bidimensional and unidimensional (maximal diameter) measurements, the resulting Response Evaluation Criteria in Solid Tumors (RECIST) guidelines simplified tumor measurement to one dimensional. The categories of the WHO conceptual model were retained and assigned functionally equivalent cut-off values to maintain historical consistency, keeping evaluation over time feasible and meaningful.
The original WHO categories of PR and PD, validated and updated by RECIST 1.0 and 1.1, still form the basis for imaging endpoints, objective response rate (ORR) and progression-free survival (PFS), which are widely used in contemporary clinical trials. However, the long reign of this conceptual model is challenged by new therapeutics which cast doubt on its biological rationale–that extensive tumor shrinkage is the only sign of drug activity. Cytostatic targeted molecular therapies provide clinical benefit without producing enough shrinkage to qualify as PR, while immune-checkpoint blockade agents can extend survival despite initial increase in tumor size.
Volumetrics: Truthful Representation of Total Tumor Burden and More Sensitive to Asymmetric Tumor Changes
Within the last few decades, multislice CT imaging and tumor segmentation software have enabled three-dimensional measurement of tumor volume. This volumetric approach provides a more accurate estimate of total tumor burden by overcoming the assumption (embedded in RECIST’s unidimensional measurements) that all tumors are spherical and respond to therapy with uniform spatial changes. As with the WHO guidelines, developing volumetric biomarkers requires establishing reproducibility, here provided by “coffee break” imaging studies. Volumetric assessment was promisingly found to have less proportional variance than unidimensional measurement, indicating that volumetrics has improved reproducibility and thus improved capability of detecting smaller, asymmetric change in tumor size over RECIST unidimensional measurement.
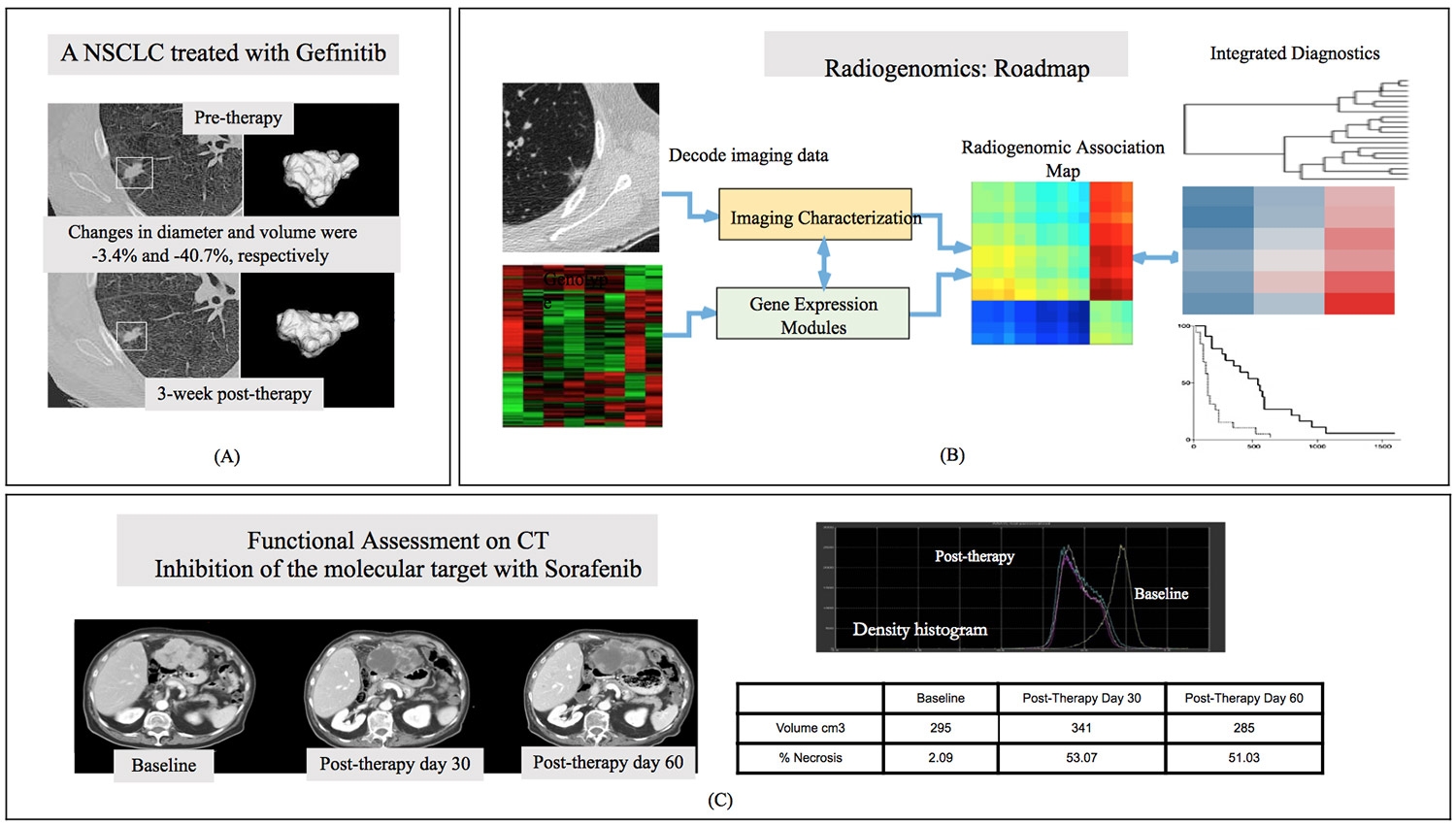
In pilot data, volumetrics outperformed unidimensional criteria in early (3-week post gefitinib targeted therapy) differentiation of wild-type NSCLC tumors from those with the sensitizing EFGR mutation (Figure 1A). Similarly, analyses of chemotherapy clinical trials in sarcoma showed that volumetric criteria for progression identified more responders, and was a better predictor of clinical benefit, than unidimensional RECIST criteria. However, other work has found no difference in response assessment between volumetric and unidimensional methods. Without volumetrics providing conclusive advantage from unidimensional methods, the field has moved on to applying the same quantitative, computationally sophisticated tools approach to study aspects of tumors other than size.
Radiomics: Mining Quantitative Information from Radiographic Images
CT images contain a wealth of detail which radiologists draw on to make visual assessments. However, such qualitative evaluations are dependent on human experience and visual perception, and thus lack reproducibility due to intra- and inter-observer variability. Quantitative image texture analysis can similarly capture information from the entire tumor and/or its microenvironment, while also yielding objective rather than subjective results based on patterns difficult for the eye to detect. This is a mature approach, introduced over four decades ago and widely used during the development of computer-aided cancer detection and diagnosis. It has recently become known as radiomics (by analogy to genomics). Indeed, advances in high-throughput mining and big data curation enabled both radiomics and genomics to promise an era of precision oncology in which an individual’s treatment is matched to their biomarker-assayed receptivity (Figure 1B).
Radiomic biomarkers have predicted a range of cancer outcomes, including (but not limited to) tumor prognosis in lung, head and neck, oropharyngeal and colorectal cancers, tumor distant metastasis in lung cancer, tumor staging in colorectal cancer, reaction to radiotherapy in esophageal cancer, tumor malignancy in lung cancer screening, and patient survival in lung cancer. Radiomics excels at assessing responses that violate the historical assumption of tumor shrinkage, such as sorafenib in advanced hepatocellular carcinoma (Figure 1C). Here, central tumor necrosis is a new response pattern that is captured qualitatively by experienced radiologists and quantitatively by radiomics as a change in image density, but would be overlooked by imaging endpoints based on size alone.
Pilot work in lung cancer showed that radiomic features measuring heterogeneity were able to predict EFGR mutation status in NSCLC tumors and were associated with gefitinib response. Such results herald a role for sophisticated quantitative analysis to not only improve previous imaging biomarkers in helping discover tissue biomarkers, but also complement genomic biomarkers using information acquired at low cost from routinely acquired CT scans that could non-invasively assess the mutational status of the entire tumor (unlike conventional biopsy) with high spatial precision (unlike liquid biopsy). A new term, radio-genomics, has been coined for this promising overlap.
Radiomics and volumetrics both require tumor contours to be annotated on the image series. For humans, this is a time-consuming process subject to inter-observer variability; fully automated approaches are successful only for relatively simple targets. Semi-automated segmentation, in which algorithms are supervised by human expertise, are more efficient and have lower variability, and result in more accurate and reproducible radiomic biomarkers. However, different segmentation algorithms are a source of variance in comparing results across institutions. Radiomics is also sensitive to imaging acquisition parameters, as demonstrated using the original coffee break study which used a range of slice thicknesses and reconstruction kernels. While radiomic features were generally reproducible across a wide range of commonly-used imaging parameters, some (especially texture features) had considerable variability when computed from images reconstructed at different imaging parameters. Additional variability studies and clinical validation are building much-needed evidence that will identify and harmonize optimal imaging acquisition parameters to ensure that radiomics has the reproducibility necessary to serve as an improved imaging biomarker.
Tumor Kinetic Modeling: Conceptualizing Tumor Progression and Response as Continuous Variables
Progress results from conceptual as well as technological advances. While tumors are no longer measured by calipers, available data remains under-utilized because contemporary response assessment sorts it into coarse categories from that same era. Modeling tumor growth and decay as continuous variables can enhance comparison between clinical trials and reveal results obscured by categorical analysis.
Kinetic modeling fits quantitative estimates of tumor burden to a curve, allowing for calculation of an exponential rate of decay and growth. Response to therapy is considered as the product of decay of the fraction of the tumor that is sensitive to treatment (initially large, driving initial shrinkage), and the growth of the resistant fraction (which eventually predominates, causing the tumor to swell again). Growth rate (g) strongly correlates with overall survival in prostate cancer, renal cell carcinoma, and metastatic breast cancer, and affords a significant increase in predicting response to therapy when added to a model of baseline variables.
Because growth rate can be analyzed even as a tumor is shrinking, kinetic modeling offers a promising tool for early prediction of the success or failure of potential therapies. Simulated sample size analysis using data from the comparator groups of large clinical trials indicates that clinical trial endpoints based on g could create a striking reduction in the number of patients required to establish treatment superiority, improving the cost-effectiveness and success rate of new cancer drug development.
Kinetic modeling has performed well using relatively coarse estimates of tumor burden, such as tumor diameter in renal cell carcinoma, but pilot work suggests that using volumetric estimates produces better fit and improves outcome prediction. Kinetic models based on radiomics may gain further power by analyzing image features which distinguish the resistant and sensitive tumor fractions, or characterize interfaces between them.
Artificial Intelligence: Machine Learning from Clinical Experience
The 1956 summer workshop on artificial intelligence (AI) held at Dartmouth College raised the curtain on modern AI. After about a half century of “winter,” AI is entering almost every corner of our daily life thanks to powerful computers, breakthrough in AI techniques including convolutional neural networks (CNN), and big data. Disease diagnoses based on radiographic images is one of the first areas in medicine that began to shine this light.
This remarkable development stems directly from RECIST’s transformative impact on the field. Big data is a key enabler of AI because the accuracy of assessments made via machine learning, as with human expertise, depends on accumulating sufficient experience. The other enabler is the unexpectedly rapid advance of CNN, an AI technique roughly modeled on the brain’s visual processing architecture. CNN algorithms are typically pre-trained on immense databases of natural images, but they need large and clinically appropriate cancer-specific imaging and clinical datasets in order to learn to identify the patterns that CNN research has proven are capable of differentiating tumors from lung nodules, wild-type tumors from mutants, or treatment responders from non-responders. These databases exist because developments like RECIST made CT so widespread and clinically useful that, although the progress of AI in imaging biomarkers currently seems amazing, this prerequisite transformation of the field by computers is taken for granted.
Using CNN to develop imaging biomarkers, in the form of algorithmic predictive models that can be reproducibly applied to quantitatively evaluate cancer datasets, is likely to have the largest initial impact on the design of clinical trial endpoints where objectivity and reproducibility are essential. Input and supervision by human experts will be critical to this process, as shown by the relative superiority of tumor segmentation algorithms which aid radiologists over fully automatic versions.
CNN and other AI techniques can also guide personalized decision-making in routine clinical practice by quickly digesting all the relevant information on individual patients, including medical history and patient-reported outcomes as well as imaging results, and automatically discovering relevant patterns that can guide the assessment of the treating oncologist. In this way, AI is neither more nor less transformative than the first written description of cancer in 1600 BC: Both permit judgements to be made on the basis of more clinical experience than can be accrued within one human lifetime, and become more useful with the growth of evidence in the public domain.

