From Data to Knowledge:
Using Semantic AI In Biomedicine
iomedical data are accumulating at an exponential rate. Knowledge derived from these data is not growing nearly as fast. There is an urgent need for technologies that can move beyond analysis of individual datasets to more effectively extract and synthesize information spread across many public and internal data sources. Semantic AI platforms combining knowledge graphs with bioinformatics, AI, and machine learning applications can provide pharmaceutical and research organizations with continuously updated data-driven knowledge, aggregating information from billions of data points to inform R&D processes and accelerate drug discovery and development.
The grand challenge for the next decade is to accelerate the translation of data to knowledge such that it can lead to actionable insights, precise therapeutic interventions, and healthcare strategies. So how exactly does data become knowledge?
Knowledge never forms in a vacuum. It derives from changing or updating our current understanding based on new observations or data. Therefore, a prerequisite for growing knowledge from data is the ability to relate new data to prior knowledge.
Hypothesis-driven research provides a natural framework for leveraging data to update knowledge. In this framework, new data are collected to address a specific question or hypothesis. Prior knowledge is used to formulate the hypothesis while at the same time providing the background necessary for interpreting new data and updating our knowledge.
In contrast, Big Data obtained through high-throughput genomics and phenotypic screening, or large collections of clinical, EMR, and real world evidence (RWE) are, more often than not, unaccompanied by precisely defined hypotheses. Combined with novel machine learning frameworks and natural language processing, these data can quickly generate thousands to billions of associations or statistical relations, yet they do not readily translate to increased knowledge. The hardship is no longer in data generation but in interpretation: What do the new relations mean? Has anyone observed similar dependencies or relations before? How do they relate to the current state of knowledge, and how do we update our knowledge based on the observed relations? What are the most promising targets or biomarkers given all of the data- and literature-driven knowledge collected to date?
These are common questions that we need to answer increasingly quickly with increasingly more data. And these questions arise not just in early discovery but throughout the drug development process. Increasingly, large and diverse datasets are generated as part of early stage clinical trials in search of biomarkers to guide patient selection and combination strategies in subsequent stages of development. This dynamic, data-rich environment requires navigating a growing space of relevant data, and relating new observations to prior knowledge to draw non-obvious connections and drive new insights.
Recent machine learning and AI technologies offer a significant hand in addressing these challenges. However, not all AI techniques are equally applicable to specific problems in the biomedical domain. In biomedicine, a unique difficulty has to do with the fact that datasets are often not as large as they are diverse, e.g., spanning different experimental technologies, conditions, and contexts. There is also a strong need for methods that are interpretable by human experts and allow for relating data-driven knowledge to existing domain knowledge and expertise.
Based on our experiences developing the Biomedical Intelligence Cloud, a powerful approach to this problem is to combine AI and machine learning with modern semantic knowledge graph technologies—an approach referred to here as semantic AI. Simply put, knowledge graphs refer to large networks of entities, their semantic types, properties, and relationships between entities. In biology and biomedicine, knowledge graphs can span genes, proteins, pathways, diseases, drugs, phenotypic and clinical outcomes, and other entities. Advanced knowledge graphs are easily updated and can capture and represent information from various sources. Thus, they can serve as continuously updated repositories of data-driven knowledge, providing a framework for highly interpretable and flexible knowledge representation. As new datasets are ingested and analyzed through appropriate bioinformatics, machine learning, or AI applications, the graph can integrate new relationships and update its knowledge representation. Such a process can be divided into three main stages: (1) semantic data integration, (2) data analysis and information extraction, and (3) information contextualization and knowledge update (Figure 1).

Figure 1. Key layers of a semantic AI system for translating data into knowledge and new discoveries. Input data and literature are semantically annotated and analyzed through a collection of bioinformatics, machine learning, and AI applications or apps. Information obtained in this way is dynamically integrated in the context of millions of prior analyses, and used to update the dynamic data-driven knowledge graph. Biomedical researchers and decision makers can use the knowledge graph to uncover hidden connections that are supported by multiple types of evidence and make data-driven decisions across the drug development pipeline. The knowledge graph can also provide prior knowledge for subsequent data-driven analyses in the analysis layer.
Stage 1: Semantic Data Integration
The first step is data integration. Notably, integrating data is more than just storing it in one place—be it an on-site, cloud-based, or a hybrid data warehouse or data lake. What is needed is deep semantic integration. Being able to capture and represent not only the entities encoded in the data but also the properties of these entities, and how those properties were measured in underlying experiments or clinical cohorts, is key to making data findable, interoperable, and reusable (see also FAIR principles). It is also a prerequisite for building an automated system for updating knowledge from data. Semantic integration facilitates convergence of evidence and information coming from multiple sources, including multidimensional omics data, phenotypic data, molecular networks and pathways, literature, clinical trials, and other structured and non-structured data that may exist both within the organization and outside in the public domain.
Stage 2: Intelligent Purpose-Built Applications to Extract Information from Data
A rich, semantically integrated and interoperable collection of data is instrumental in fueling statistical and machine learning engines that can extract meaningful information from these data. Biopharmaceutical research and development requires a growing range of tools to support analytics applications throughout the drug development pipeline from early target discovery and validation, to mechanism of action and efficacy studies, to patient selection and developing companion diagnostics. A modular app-based approach to tool development is recommended as it allows for easily incorporating new methods and modeling approaches as they become available. Specialized applications and models implementing methods from bioinformatics, systems biology, NLP, AI, and machine learning can be purpose-built and trained to support specific R&D tasks.
Stage 3: Combining and Contextualizing Information to Update Knowledge
As new datasets are integrated and analyzed by the appropriate apps, the information being generated can be captured in the form of knowledge graph relations and dynamically integrated with prior knowledge. With access to the combined knowledge from semantically-linked data and literature, a user can quickly identify a biomarker or a target candidate that is supported not just but a single data source but by tens of preclinical and clinical datasets and/or literature evidence. While information coming from each source may be noisy, incomplete, and susceptible to biological and experimental biases, these problems are unlikely to reproduce in the same exact way across many independent sources. This allows specialized knowledge graphs to boost signal over noise and assign higher confidence to findings which are observed across multiple studies. Importantly, apart from directly informing human analysts, knowledge aggregated in the knowledge graph can also feed back into the machine learning applications to provide prior knowledge for subsequent data analyses.
A Clinical Trial in the Context of Prior Knowledge
To illustrate the above concepts with a timely example, let’s look at a case study focused on identifying biomarkers predicting response and resistance to immune checkpoint inhibitors presented a few months ago at the Precision Medicine World Conference (PMWC 2019). A study published last year in Nature made data available from the IMvigor210 clinical trial of the PD-L1 inhibitor atezolizumab in urinary bladder cancer patients. Pre-treatment somatic mutation, gene expression, pathology measurements, and associated clinical outcomes were collected with the goal of identifying biomarkers of response and resistance and, for non-responding subpopulations, identify potential combination strategies that may help overcome single-agent resistance.
How can prior biomedical knowledge (e.g., captured in the form of a knowledge graph) help inform AI and machine learning approaches in the analysis and interpretation of such a rich multidimensional clinical trial dataset? As it turns out, it can do so in a number of different ways (Figure 2).
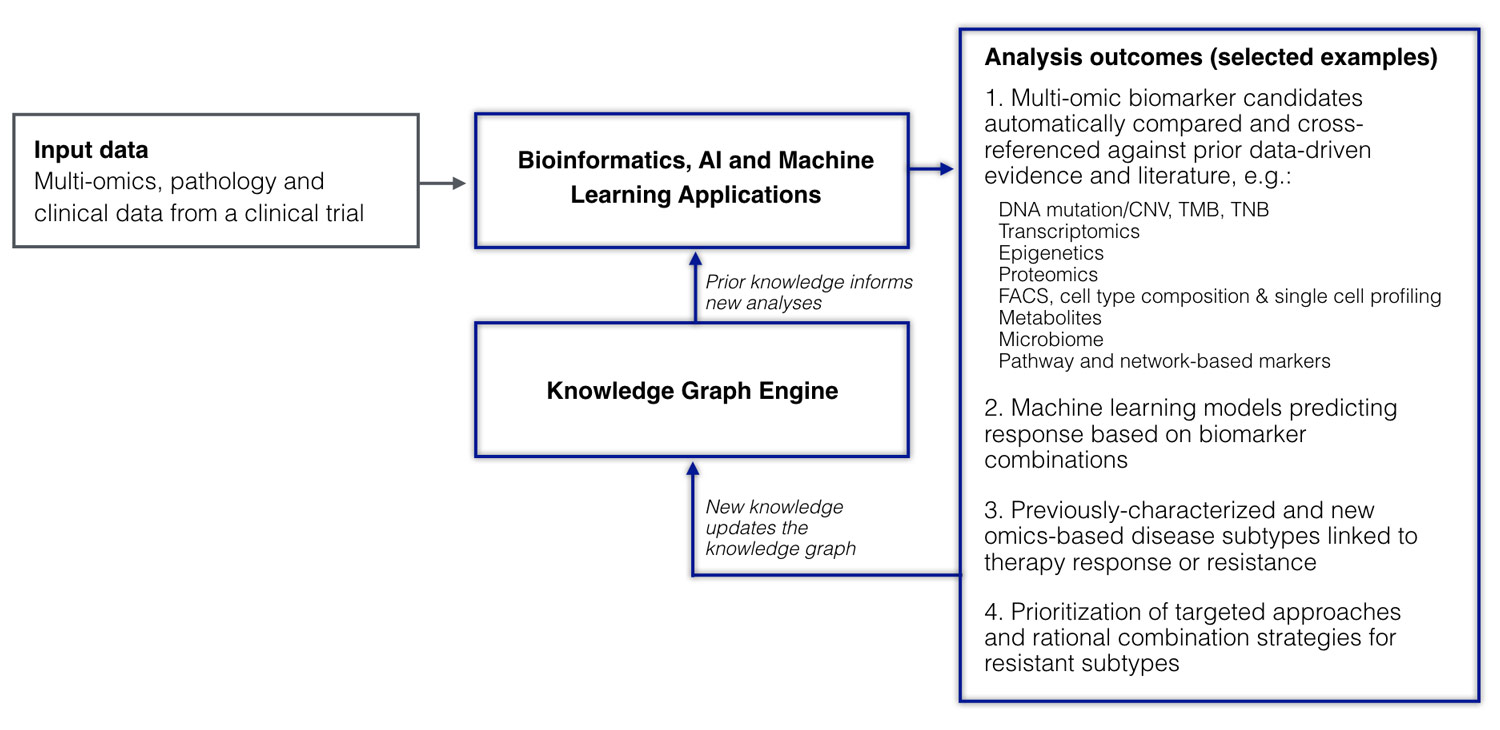
Figure 2. A clinical trial dataset analyzed in the context of the knowledge graph. Multiple systems biology and machine learning applications are used to analyze multidimensional omics, pathology, and clinical data uncovering pathways, building predictive models, and discovering disease subtypes associated with mechanisms of response and resistance. Prior knowledge from the knowledge graph is used to inform the analysis and make predictions; new knowledge resulting from the analysis of the new clinical trial cohort is used to update the knowledge graph.
First, prior knowledge of molecular pathways can be instrumental in identifying meaningful and interpretable biomarkers from high-throughput data. In our case study, a system equipped with over 20 molecular pathway and network domains spanning over 100,000 functional components identified TGFβ as one of the very top pathways associated with atezolizumab resistance, recapitulating the earlier finding and doing so without any input from a human expert. Furthermore, pre-integrated knowledge from prior studies allowed the system to identify other conditions and biological contexts in which the same pathway biomarker showed relevance. In this way, we were able to automatically identify up to ten additional cohorts in which TGFβ pathway expression was associated with clinical outcomes.
Moving beyond individual markers, a semantic AI system can leverage multidimensional patient data from the trial to build machine learning models that identify optimal combinations of features predicting response. Such a built model using the IMvigor210 data identified high tumor mutation and tumor neoantigen burden in combination with expression of the WNT signaling pathway as the most important features for predicting response. WNT signaling has previously been shown in other histologies to be associated with lack of immune cell infiltration, which may negatively impact IO therapy response—knowledge that can also be captured by prior data- and literature-driven evidence in the knowledge graph. Notably, while individual predictors such as WNT pathway expression might not always be significant on their own, they may become important in combination with other predictors.
Finally, by utilizing prior knowledge of molecular cancer subtypes, a semantic AI system can identify and, in some cases, predict subtypes associated with drug response or resistance. In the case of the IMvigor210 data, the system used in our case study quickly recapitulated a previously characterized association between the genomically unstable subtype of bladder cancer and partial response, but also found a new molecular subtype not previously reported that showed association with complete response in the IMvigor210 trial.
Prior data- and literature-driven knowledge (including knowledge of molecular pathways, disease subtypes, and results from previous patient cohorts) can boost the analysis of other trials across different cancer histologies as well as non-oncology indications. Recently, at the 2019 Annual Meeting of the American Association for Cancer Research (AACR), we presented a new resource called the Cancer Subtype Ontology (CSO) which spans over 800 previously published and internally developed cancer subtypes across over 35 cancer histologies. Subtypes in the CSO are linked to key genomic, transcriptional, and epigenetic modifications at the gene and pathways level and annotated with additional features such as tumor mutation burden and tumor-immune infiltration profiles as well as response and clinical outcomes data from prior cohorts. We used the CSO to analyze 19 published cohorts treated with immune checkpoint inhibitors and make predictions about clinical trial outcomes in the context of CSO subtypes. We found that for the majority of the trial cohorts, specific subtypes existed in the CSO which were associated with clinical outcomes such as therapy response and/or resistance and survival times.
While the above examples represent retrospective analyses, similar strategies are being applied to make prospective predictions for new cohorts. The results and evaluation of such predictions can be integrated back into the knowledge graph (Figure 2), providing a framework for forward translation of data-driven knowledge to inform clinical trials as well as reverse translation of findings from clinical trial analyses to inform the next steps in the clinical development process.

