Translational Science
Advancing Cancer Immunotherapy with Proteomics


Introduction by Gary J. Kelloff and David Parkinson, DIA Global Forum Translational Science Content Editors
ver the past decade, successes in precision medicine have been built on the backbone of scientific advances in genomics. However, more than good genomic science is needed. In fact, understanding the effects genomic alterations have on cell function is equally or more important.
One way of measuring this functionality is evaluation of genome-driven protein expression (proteogenomics), but this effort has been hampered by irreproducible, non-quantifiable assays and a lack of clinically practical assays. Two efforts to address these challenges are described in this month’s translational science overview:
- The National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium’s program to develop robust multiplex protein assays using multiple reaction monitoring (MRM) mass spectroscopy, which measures multiple immunomodulatory factors simultaneously
- The use of nucleic acid programmable protein arrays (NAPPA) technology to evaluate the effects of therapy on protein expression in individual patients treated with immunotherapy, an effort that is pertinent to predictive medicine in oncology and currently a high priority in clinical cancer research.
Issues at Hand
Immuno-oncology (I-O) is an emerging field revolutionizing the use of immunotherapy to treat malignancies. Despite widespread enthusiasm for immunotherapies in oncology, the field faces challenges surrounding immunomodulation in cancer, especially for solid tumors. A medical imperative exists for us to understand and quantify the many immuno-modulatory factors in the cancer-immunity cycle, as these factors may be viable therapeutic targets or may influence antitumor responses to immunotherapy. Most notably, programmed cell death ligand-1 (PD L1) is currently the only tissue-based biomarker, and there are no blood-based soluble biomarkers to select patients for immune therapies. Significant subsets of patients do not respond to immunotherapy, and others suffer treatment-limiting adverse events. Therefore, there is a need to understand factors that both allow and prevent a patient’s response to immunotherapy and to identify biomarkers that can predict response as well as help manage autoimmune toxicity. Providing multiplexed assays that enable quantification of many immunomodulatory proteins beyond PD-L1 in both the tumor microenvironment and periphery should help determine a patient’s suitability for monotherapy with checkpoint inhibitors. Additionally, these multiplexed assays could identify patients who may require combination I-O treatments to increase the likelihood of response and improve cancer patient management.
Protein Expression Monitoring
Existing immunoassay methods have limitations for quantifying proteins and are difficult to develop. For example, immunohistochemistry (IHC) assays are semi-quantitative and difficult to standardize and multiplex, whereas Western blots are neither quantitative nor multiplexable, and are fraught with interferences. Each of the four IHC-based assays to PD-L1 is associated with a different therapy. Because this represents a unique challenge to medical professionals and patients, a cross-industry working group among the US Food and Drug Administration (FDA), the American Association for Cancer Research (AACR), and American Society of Clinical Oncology (ASCO) developed the Blueprint Initiative to review the issue of harmonization between the PD-L1 assays. Having a broadly available, standardized I-O assay panel would help address the need for harmonization.
Some investigators have turned to mRNA-based measurements to infer protein expression of immuno-modulatory factors (e.g., Nanostring) and circumvent the technological limitations of conventional immunoassay platforms. However, for human breast, colorectal, and ovarian cancers, RNA levels have shown to be unreliable predictors of protein levels or activity. Thus, direct quantification of proteins is desirable. The National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) of the Office of Cancer Clinical Proteomics Research and the Paulovich Laboratory at the Fred Hutchinson Cancer Research Center partnered to develop a multiplex assay panel that quantifies immunomodulatory proteins in the tumor microenvironment and/or in the periphery. A committee involving I-O experts from the pharmaceutical industry and academia selected the protein targets to be included in the multiplex assay panel.
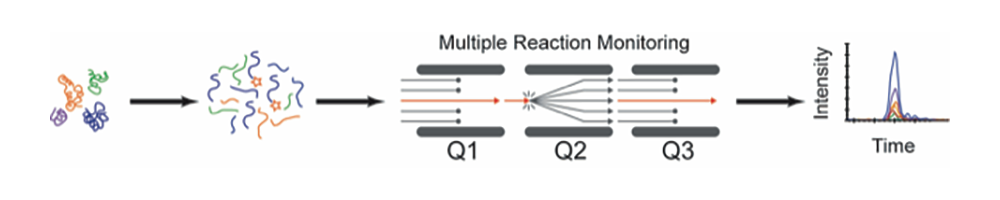
MRM assays
Proteins are digested with trypsin and proteotypic peptides are isolated for multiple reaction monitoring mass spectrometry in a triple quadrupole mass spectrometer. A stable, isotope-labeled version of each peptide is spiked as an internal standard, enabling precise relative quantification of the target across biospecimens. MRM-based assays are highly specific and precise and can be readily multiplexed and standardized across laboratories.
The multiplex assay will be based on peptide immuno-enrichment coupled to targeted, multiple reaction monitoring mass spectrometry (immuno-MRM-MS). This next generation protein quantification platform is a highly attractive alternative to current immunoassay platforms given its speed, high specificity, quantitative precision, and ability to quantify panels of proteins in a multiplex manner. Furthermore, the use of internal isotopic standards means that MRM-based assays can be standardized and harmonized across laboratories. Immuno-MRM assay panels can be used in almost every setting where one would typically use Western blotting, ELISA, or IHC assays. The exception are cases which require single cell analysis or geographical localization, in which case IHC or flow cytometry is necessary. Thus, the I-O immuno-MRM assay panel will be highly complementary to existing immunoassays for quantifying I-O proteins and further help the oncology community answer why immunotherapy does not yet work for all patients or cancers.
The I-O multiplex immuno-MRM-MS panel will be characterized in compliance with the CPTAC’s established guidelines for MRM-based assay bioanalytical validation. All assay protocols and monoclonal antibodies will be made broadly available to the research community via NCI’s open source portals (www.assays.cancer.gov and www.antibodies.cancer.gov). The I-O multiplex immuno-MRM-MS panel will impact clinical projects by enabling comprehensive quantification of immunomodulatory proteins in the tumor microenvironment and periphery, potentially guiding a patient’s selection for either mono- or combination immunotherapies.
Convergence of the Cellular and Humoral Immune Systems
As shown by numerous studies, cell-mediated immunity plays a crucial role in the response to immunotherapy. However, humoral immunity, including coordination between T and B cells during the anti-tumor response, remains largely unexplored. B cells likely affect therapeutic response through diverse and relevant immunologic functions such as antigen presentation, immuno-stimulatory cytokine production, and possible direct tumor lysis via anti-tumor antibody responses. Given the active immune modulation that occurs during checkpoint immunotherapies, the antibody response represents a promising, yet so far fairly unexplored, avenue of research that may shed light on the biological mechanism of response to immunotherapy. The specific targets and amplitude of pre-treatment humoral immunity may predict a patient’s overall anti-cancer response. Moreover, the humoral immune system could also be an early indicator of immune-related adverse events (irAEs) that are unleashed by immune checkpoint immunotherapy, some of which are likely antibody-mediated and can be treatment limiting. However, none of these related characteristics—the antigens targeted, their functional role, and their use as biomarkers of immune responses—have been systematically studied.
Protein microarrays provide a multiplexed, high-throughput platform to profile global antibody immune responses against thousands of proteins in parallel without the display biases that are inherent to library-based methods, such as SEREX and phage display. Different classes of antibodies can be detected simultaneously using secondary antibodies labelled with different fluorophores. The LaBaer laboratory at Arizona State University (ASU) pioneered and developed an innovative protein array platform termed nucleic acid programmable protein arrays (NAPPA). NAPPA entails programming cell free protein expression extracts with cDNAs to express and immobilize the full-length proteins in situ at the time of the assay without the need for advanced protein purification. The chemistry behind NAPPA is advantageous because it encourages human proteins expressed in a human milieu, including chaperone proteins, to fold naturally. The “just-in-time” production abrogates concerns about protein stability during storage, because “fresh” proteins are made for each assay.
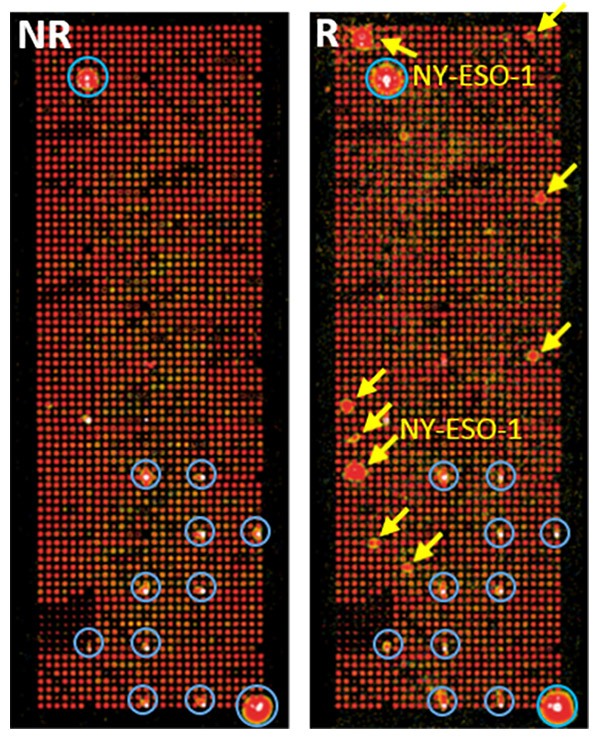
NAPPA for detection of antibody responses in immunotherapy responders
NAPPA arrays are probed with pooled samples from melanoma patients treated with anti-PD1 therapy. Red arrows indicate strong differential immune responses between responders (R) and non-responders (NR). Yellow circles indicate positive control antigens.
The plasmid repository at ASU is on its way to having the complete complement of protein-encoding human genes (DNASU.org). NAPPA will enable the interrogation of comprehensive antibody profiles against the full human proteome in patients before and after immunotherapies. Emerging data indicate that the level of the tumor non-synonymous mutational burden correlates with response to checkpoint immunotherapy, supporting the hypothesis that mutated neoantigens are an important immune target in responding patients. The flexibility of the NAPPA platform makes it extremely convenient to produce protein arrays with any cDNA sequences. Thus, NAPPA can be used to study the patient’s antibody responses to mutated proteins encoded by their own tumor tissues on “personalized mutated neoantigenome” arrays.
Antibody profiles can be analyzed with clinical information, such as treatment response or autoimmune toxicity. Robust, large-scale investigations into plasma antibody profiles and its associated changes with immunotherapy may lead to an antibody biomarker panel that can be paired with other modalities and help patients select the most appropriate immunotherapy with manageable irAEs. Understanding the mutation status of antigens that are targeted by checkpoint immunotherapies could be used to develop the next generation of tumor immunotherapies with personalized peptide vaccines, or to expand neoantigen-specific T cells ex vivo for transfer to patients, both of which could be combined with checkpoint therapy.
Overall Conclusion
In summary, despite the major successes we have observed using immunotherapies to treat cancer, finding tools to identify patients who will benefit the most from these treatments and those who are likely to suffer adverse events remains a challenge. As proteomics has matured, combining precision with scalability, it now offers two exciting complementary approaches.
First, a quantitative, multiplexed approach for measuring the levels of various immunomodulatory factors through immune-MRM MS will lead to biomarkers that predict these outcomes.
Second, a global examination of the humoral immune response using proteome-scale and personalized protein microarrays will shed light on the coordinated immune response as well as the specific targets of the immune response. Combining these tools may lead to personalized diagnostics that help physicians manage immunotherapy and new adjunctive therapies, such as personalized vaccines.
About the Authors

Ji Qui
Research Associate Professor,
Center for Personalized Diagnostics
Biodesign Institute
Arizona State University

Joshua LaBaer
Professor, School of Molecular Science
Director, Center for Personalized Diagnostics
Executive Director, Biodesign Institute
Arizona State University

Henry Rodriguez
Director, Office of Cancer Clinical Proteomics Research,
National Cancer Institute, NIH

Amanda G. Paulovich
Member, Clinical Research Division
Director, Early Detection Initiative
Fred Hutchinson Cancer Research Center
Member, Fred Hutchinson/University of Washington Cancer Consortium
Professor, Department of Medicine/Division of Oncology
University of Washington
Associate Faculty Member, Molecular and Cellular Biology Program
University of Washington